Whole-cell models, or computational models that account for the integrated function of every gene and molecule in a cell, have been described as “the ultimate goal” of systems biology, and “a grand challenge for the 21st century”. The cell simulations captured a wide range of cellular activities, and suggested future experiments that were validated experimentally.

On a biological level, our bodies are made up of many biological components that are integrated at and communicate on multiple scales, multiple languages. From our genome to the molecules and cells that make up the organs in our bodies all the way out to ourselves in our world, the drugs we take, the disease we develop: we are fundamentally built upon a communication network. Complex interactions in this multi-language biological system are leading to emergent properties, properties of cells, tissues and organisms to functioning as a system.
CellMo embarks on the complex task to create a holistic cell model capturing complex cell dynamics and context (i.e. tissue, disease) specific states.
CellMo combines state-of-the-art network analysis methods with revolutionary deep learning techniques, which are also responsible for the breakthrough successes achieved in large language models in recent years (e.g. BERT, GPT3 in translation, chatbots, etc.). These methods make it possible to manage biological processes as the descriptive language of biology and to analyze complex molecular and pathophysiological relationships more precisely and in greater depth than ever before.
As an example, CellMo can be used to “translate” the cellular effects of a disease into the pathophysiological effects of a drug to discover low development cost, repurposed drug compounds for a disease.



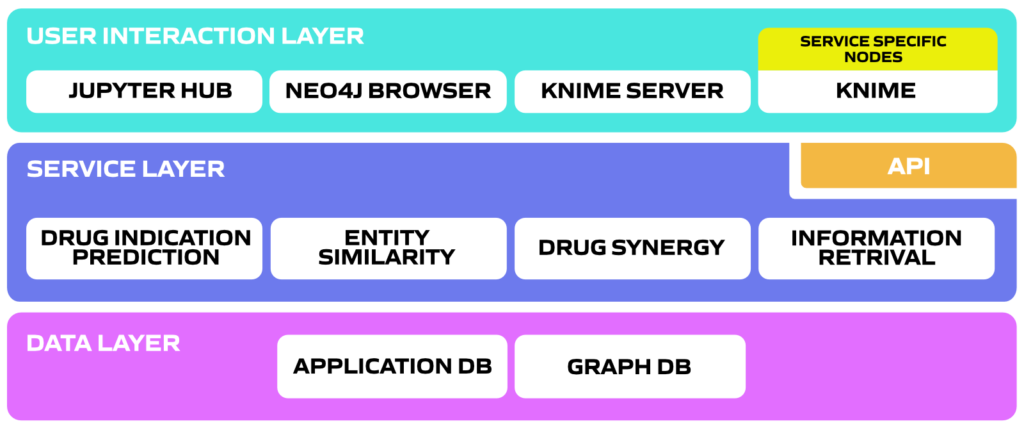
It provides an integrated network of biological concepts and relations for users to create complex systemic level biological queries and for algorithms to identify unknown holistic relations. The integrative network database assembled from 29 different databases of genes, compounds, diseases, and more. The network contains 47,031 vertices with 11 different labels and 2,250,197 edges with 24 different labels.
Biological questions can often be answered by looking up certain type of paths in the network. E.g. looking up compound-treats-disease paths would answer which compounds treat a disease, or disease-upregulates-gene-participates-molecular function paths can indicate in which diseases are genes -encoding enzymes/proteins with a certain molecular function- upregulated. Such sequences of vertices following this specific labeled path are called ’walks’ in the network.
Data analytics services that help researchers to understand the data and explain relations, to analyse the data and draw conclusions, and also, to find new possible relations between entities e.g. predict new indications for a drug, find novel, alternative treatments for a disease or uncover probable synergistic relations between drugs.
User interaction layer allows for user to access the CellMo platform, which is available on various open source interfaces. Using the CellMo toolset domain experts can create complex drug discovery workflows without deep technical skills, while being compatible with a large ecosystem of cheminformatics tools.

Target-based drug discovery and repositioning is still largely manual work today. The value of drug repositioning lies in the fact: using the approved active ingredient or drug component for new purposes, it is not necessary to carry out many steps of the mechanism of action investigation and approval. Thus, the average 12-15 years of drug research time can be radically reduced to 4-6 Years duration. Many existing computational platforms and databases exist and are used for integrative analysis, but there is no broadly available tool to support systematic drug-disease prioritization. RepoX is a decision supporting platform to solve the challenge of drug repositioning/repurposing with exclusive toolset and created not just to technical users but also available as an analytics platform.
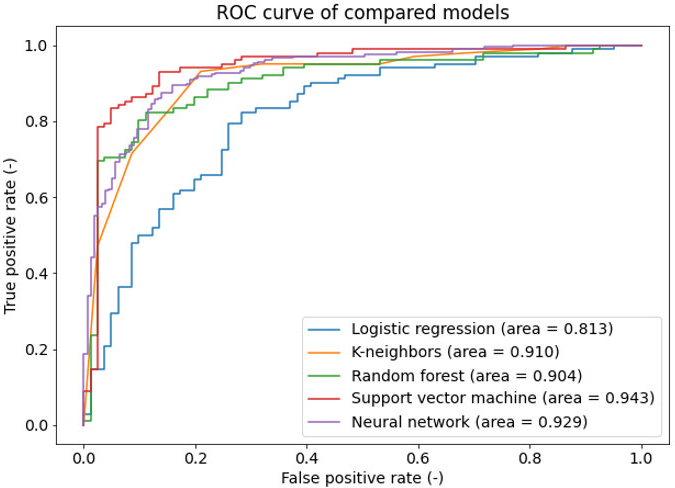
Infections caused by multi-drug resistant pathogens due to ineffective medicines could become one of the leading causes of human death by 2050, overtaking cancer as the No 1 leading cause of death, according to WHO estimates. There are many different livestock farms around the world, and keeping these crowded animal farms healthy is often difficult. Unfortunately, frequent antibiotic usage in livestock farming is a common practice, and often results in unnecessary doses of antibiotics. They are partially or entirely absorbed by the food products, including human food, contributing to the development of antibiotic resistance. Inappropriately applied antibiotics do not kill all pathogens, additionally, the more potent ones are even able to multiply, unfortunately often become resistant to the original drug. The three-year project ’Development Of Anti-Bacterial Resistance Product In Veterinary Medicine, Development Of A Supporting Response Platform’ started on the 1st of October 2021. It was launched in a consortium of the University of Veterinary Medicine Budapest and E-Group, with the support of the National Research, Development and Innovation Office.
The project aims to create an innovative drug repositioning platform for animal health. Within the project framework, E-Group will provide technological and data infrastructure expertise, including, data engineering and data science. The platform’s operation will be similar to the highly successful InnoHealth DataLake. The drug repositioning platform for animal health will be able to integrate and perform complex analysis of animal health and their (primarly) contagious diseases, veterinary public health, food safety and human health data from different sources. The platform can be used to identify multiple candidate agents to combat veterinary antibiotic resistance. As part of the One Health concept, the platform could also form the basis for a food chain safety data platform to be developed in the future.